In the Integrated Workshop (IW), students…
- are introduced to methods of modern integrative science
- become familiar with a range of quantitative methodologies and tools
- develop a close working relationship and peer tutoring amongst each other
- meet others in the PEB community
Students from a variety of academic backgrounds work together on hands-on projects and presentations that are organized into multi-week-long modules. Each module is co-taught by PEB faculty who have complimentary expertise (i.e. one from biology, one from physics or engineering, one is a theoretician and one is an experimentalist). Each module also incorporates lectures given by faculty to introduce the research topics and quantitative methods. The modules are devised so that a range of skills are acquired, and students learn from each other. Topics explored by current modules:
- “Working with MATLAB: a short tutorial” (TAs)
- “Cell Crawling” (Mike Murrell – BME)
- “Melanoma Tumor Development” (Corey O’Hern – MEMS – with Marcus Bosenberg – Pathology and Dermatology)
- “Cell Membranes” (iPoLS faculty Ben Machta – Dept. of Physics)
- “Spinal Column Development” (Scott Holley – MCDB– with Corey O’Hern – MEMS)
- “Computational Protein Design” (Mark Gerstein – MB&B– with Corey O’Hern – MEMS)
- “Microstructure of Plant Leaves” (Madhu Venkadesan – MEMS– with Corey O’Hern – MEMS)
- “Spatiotemporal Dynamics of Chromatin” (Simon Mochrie – Physics– with Megan King – Cell Biology)
- “Charge Transport in Bacteria” (Nikhil Malvankar - MB&B)
- “Spatial Organization and Crowding in the Bacterial Cytoplasm” (Christine Jacobs-Wagner – Microbial Pathogenesis – with Corey O’Hern – MEMS)
In the past, the IW consisted of laboratory modules. Below, we give a brief description of modules that had been offered in that format.
Regulated SNARE Zippering Revealed by Single-Molecule Force Spectroscopy (Y. Zhang lab)
Students study folding of single synaptic or vacuolar SNARE complex alone or in the presence of myricetin using our high-resolution optical tweezers. Myricetin is an antioxidant compound found in vegetables, fruits, and red wine. Recently, it has been found that myricetin binds SNARE complexes and likely stabilizes the SNARE complexes in a half-zippered state. The students also learn to run optical tweezers, analyze single-molecule trajectories, and identify the effects of myricetin on SNARE zippering.
Analysis of Cell Interactions in Bacterial Colonies and Model Tissues (Levchenko lab)
This module is aimed at using novel approaches (mice-and mess-fluidic chips, computational modeling, molecular biology analysis, etc.) in the analysis of cell-cell communication in complex biological settings. One system is bacterial quorum sensing, with the target of the project being to conduct the analysis in a super-high throughput and resolution fashion of the quorum behavior in E. coli, focusing on the fundamental questions underlying this collective response and implications for synthetic biology and medicine. The second system is interaction between cells in growing model blood vessels, focusing on the role of model immune responses in this system, and the use of 3D printing and other approaches in building state of the art tissue constructs. Students also get introduced to other cutting edge projects at Levchenko lab and Yale Systems Biology Institute.
Bacterial Chemotaxis (Emonet lab)
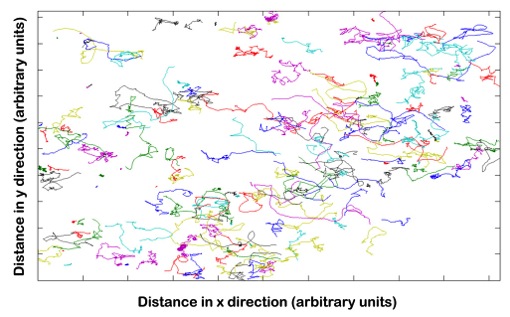
Students record the movements of wild type and mutant E.coli under various experimental conditions using microscopes. They track cellular motion and analyze the data and extract statistics on cellular motion, such as run length, run time, and tumble angles and bias, using MATLAB. They also compare their data to simulations.
 |
 |
| Bacterial trajectory plots with each trace representing a different bacterium. | Students working together to first record short movies of bacterial trajectories, and then to analyze the data. |
Single Molecule Super Resolution Microscopy: Sample Preparation to Data Post-Processing (Bewersdorf lab)
Super-resolution fluorescence microscopy has recently overcome the century-old diffraction limit in light microscopy and thereby allows unprecedented observations of sub-cellular structures. This IW module introduces students to the field of single molecule stochastic switching super-resolution microscopy. They learn how to prepare cell biological samples, how to image them and how to process the data to generate super-resolution images. Their research project involves the systematic test of key parameters of this novel imaging method to optimize performance. For example, one year, students imaged fluorescently labeled microtubules of COS7 cells to determine the excitation laser power and frame rate that would optimize the average photon count per fluorophore localization.
 |
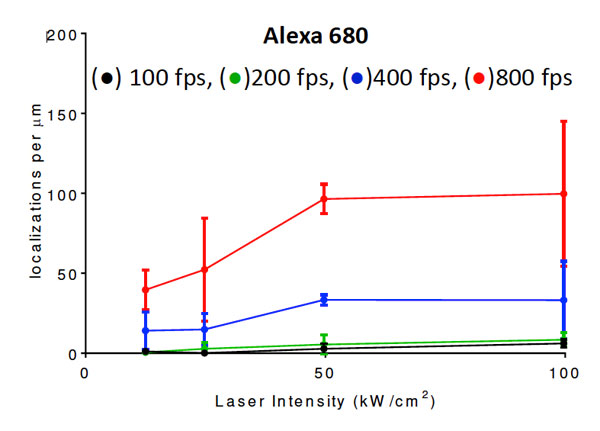 |
| A student pair working together to set up an experiment. | Localization density of a dye that was tested for high speed super resolution imaging. |
Forces Driving Cellular Motion: Skin Mechanics (Dufresne and Forscher labs)
Students receive a series of lectures aimed at providing a practical background for approaching the problem at hand. For example, students using light microscopy to study cellular mechanics are given lectures on microscope optics and digital imaging instrumentation followed by computational approaches to particle tracking. This information will then be used in experiments, where students assess traction forces used to drive cell movement by measuring the displacement of microscopic fluorescent beads embedded in an elastic growth substrate.
 |
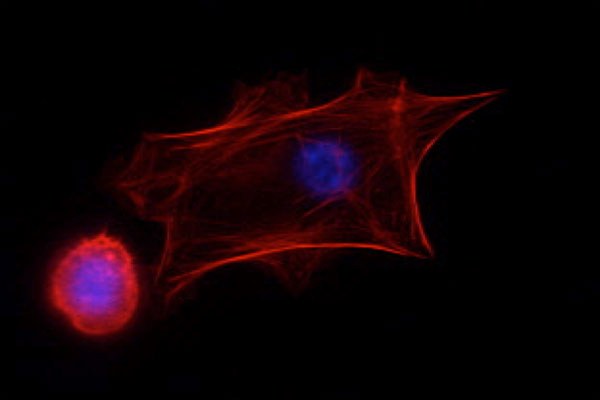 |
| Students working with TAs to familiarize themselves with various equipment. | Fluorescence image of keratinocytes; actin (red), DNA (blue) |
Analysis and Design of Protein Structures and Interfaces (O’Hern and Regan labs)
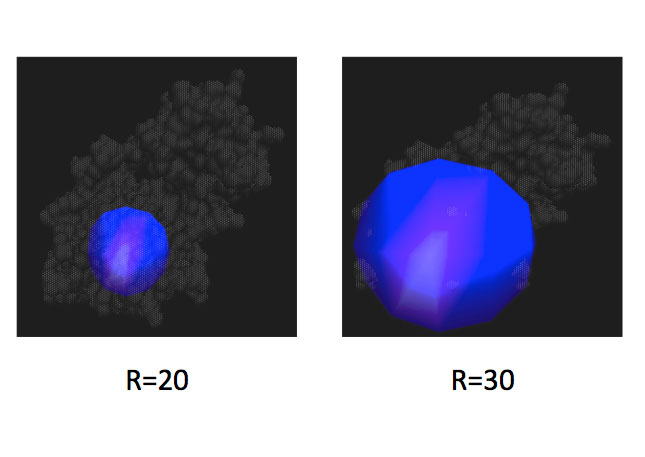
Two long-standing challenges in the fields of molecular and computational biology are the accurate prediction of protein folding, structures and protein-protein interactions. This module is focused on using computational tools and theory to explore protein structures and interfaces. One year, students examined several structural parameters from PDB files to assess the quality of the structures and their binding capability to a particular protein. During the process they also familiarize themselves with the software MolProbity. Another year, they examined the impact of steric constraints on protein core residues using coarse-grain molecular dynamics simulations, and T4 lysozyme as their model protein.
 |
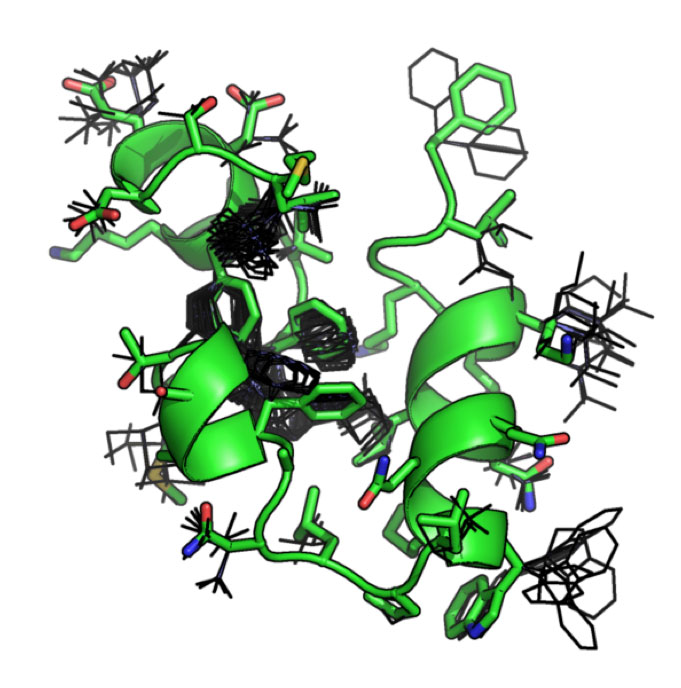 |
| Two examples of different size spheres (20 and 30 Angstroms in radius) centered on a mutated residue of interest in T4 lysozyme. The molecular dynamics simulations conducted in the module were restricted to these specific size spheres. | This image illustrates possibilities for various rotamers (low energy side chain conformations). Correct rotamer assignment may serve as a useful characteristic in assessing the quality of structures and their binding capabilities to protein partners. |
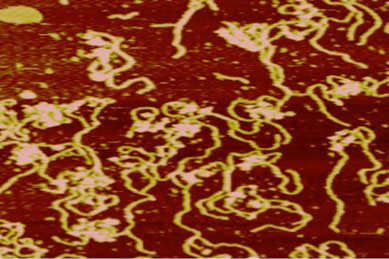
Single Molecule Experiments: Measurements and Data Analysis (Regan and Mochrie labs)
In this module, students have become familiar with conducting and analyzing a single molecule experiment. In the past, an optical tweezer setup and atomic force microscopy were used. Students spend some time prepping samples and collecting data, and the remaining time analyzing their data. For example, for the optical tweezer experiments, students purified various DNA constructs and histone proteins, then assembled nucleosomes. They then made single molecule optical tweezers force versus extension measurements on the condensed DNA and explained their data using principles from statistical mechanics and polymer physics.
 |
 |
| Atomic force microscope image of nucleosomes in the presence of Mg2+ and glutaraldehyde. | Students working together to calibrate the atomic force microscope. |
 |
 |
| Using the optical tweezer setup. | Making a flow cell used to prepare the samples. |
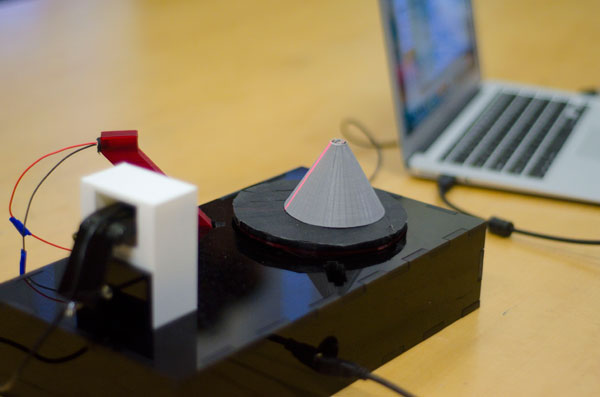
3D Printing: Design and Fabrication (hosted in the CEID)
Students created various designs with biological applications, using 3D printing and other tools available in Yale’s Center for Engineering Innovation and Design. The main projects are listed below.
Students built a 3D scanner that can be used to convert the shape of a real object into digital structure. They also wrote the Matlab program to record image-capture videos, while rotating the stage with the object to be scanned.
Students created a mini benchtop centrifuge which is capable of spinning down samples in eppendorf tubes. Their design was modular, for example, to allow for the use of different rotors.
Students made a model showing the components and how they come together in engineering protein bio-gels.
Students created the first version of a car and track that will be used to teach a physics module in Breakthrough New Haven. The module will consist of different tracks and a modular car, so that various physical principles could be tested.
 |
 |
| 3D scanner: in this case, the grey cone is being scanned. The red line is the laser as it hits the sample. The camera is capturing the image of the laser shining on the object as it rotates. | Parts of the protein bio-gel model, which can be assembled to show the interactions among the various building blocks. This model was used in an outreach event geared towards schoolchildren. MORE |
 |
 |
| Car and track design, version 1. | Mini benchtop centrifuge. |
 |
 |
| PEB students discussing their designs with Connecticut College undergraduates interested in science. | Students showing each other their designs. |
 |
 |
| Pairs working together, and with the TAs, to come up with the first drafts of their designs. | CEID staff showing students various materials and technologies available to them. For example, students could 3D print a mold to use with a machine that is able to thermoform a plastic sheet. |
Managing Oxygen in Single Molecule Biochemistry (Ganim lab)
This module focuses on minimizing the effects of oxygen damage in single molecule fluorescence and force experiments. Students learn the fundamentals of redox chemistry, and sample construction, data acquisition, and data analysis for optical tweezers force experiments.
 |
|
| Preparing a chemical reaction in an oxygen-free environment. |
Top: Working in a glovebox to eliminate the presence of oxygen. Bottom: Colorimetric assay used to monitor oxidation of sample. |
Optimization of a Microfluidic Device for Single-Cell Trapping Without Surface Modifications (Miller-Jensen lab)
Students test microfluidic devices made in the lab for various imaging applications. They learn to work with live cells, fabricate devices in a clean hood, and run experiments using them. They are also able to make recommendations for how to improve the device.
 |
 |
| A physics and a BBSB PEB student working together, with their postdoc mentor. | Physics PEB student working in a cell culture hood, getting ready for an experiment. |
 |
 |
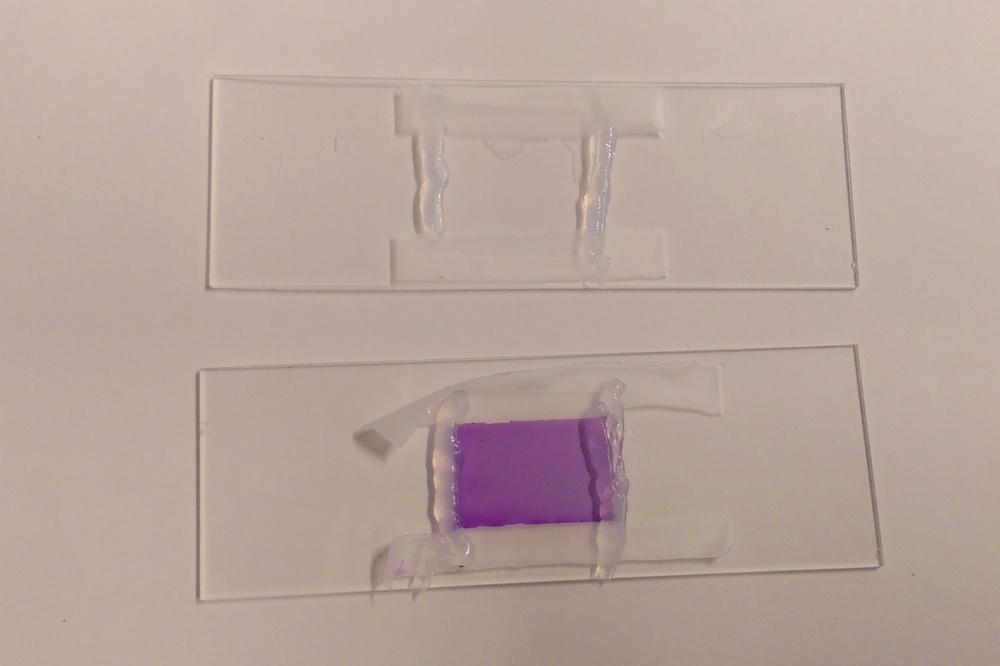
| Microfluidic workstation. | Cleaning the PDMS using tape (it’s the best way!). |
Estimation of the Turnover of Endocytic Proteins Using a Single Molecule Strategy (Berro and Baddeley labs)
Students learn about single molecule imaging and labeling techniques, working in live yeast cells. The ultimate goal is to estimate the residence time of proteins during clatherin-mediated endocytosis.
 |
|
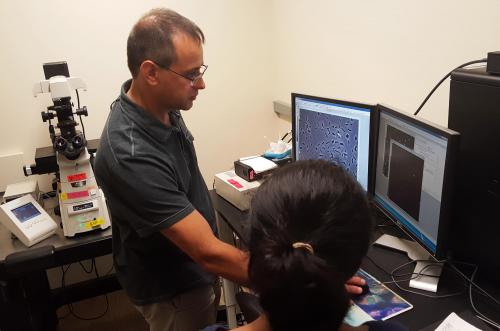
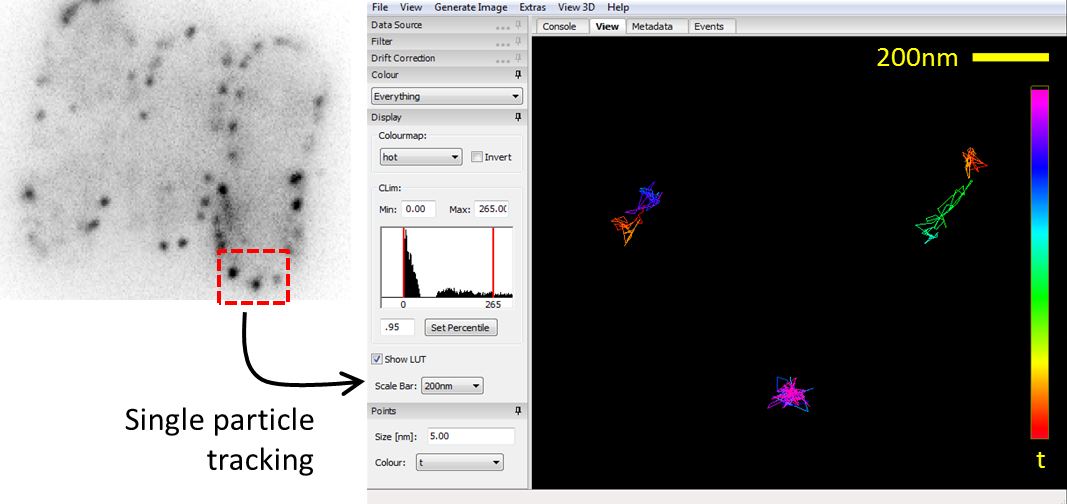
| BBSB and Physics PEB student working together to obtain data. | Data analysis, showing (top) how the trajectories of single molecules are tracked. |
Multi-Scale Modeling of Biological Systems (Campbell lab)
Students are introduced to software packages, such as MATLAB, Amber, and CHARMM, which enable simulation of biological processes across scales, from molecules to organs. They work with Prof. Campbell to identify a small project that they could work on. It can either be something that the Campbell lab is working on, or something that the student is interested in and often, a system the student already has familiarity with. A large component of this project is to do literature searches and to learn to evaluate what data could be used in models built to explore complex systems.
 |
 |
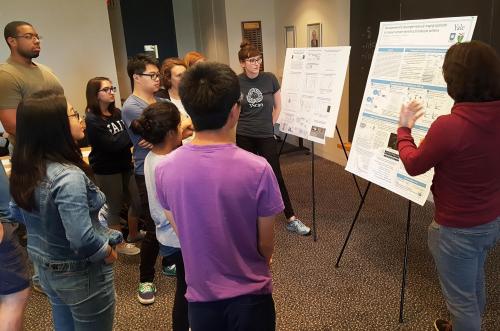
| PEB students and Prof. Campbell listening to the presentation. | A student pair presenting Modeling the effects of mutations in cardiac troponin in Familial Hypertrophic Cardiomyopathy, a project centered on the research interests of the Campbell lab. |
 |
 |
| A Physics and a BBSB student presenting on a topic of their own choosing, microtubule polymerization. | A lively discussion follows the final presentations. Prof. Campbell gives feedback on the projects and students describe their thoughts on potential future work. |
-
- 3D scanner
- Mini benchtop centrifuge
- Protein model
- Car and track